A Practical Guide to Scanning and Transmission Electron Microscopy Simulations
Contents
STEM Simulation and Post-Processing
import ase
import abtem
import matplotlib.pyplot as plt
atoms = ase.io.read("data/sto_lto.cif")
potential = abtem.Potential(atoms, sampling=0.05, slice_thickness=2)
probe = abtem.Probe(energy=150e3, defocus=50, semiangle_cutoff=20, Cs=0.0)
probe.grid.match(potential)
scan = abtem.GridScan(
start=(0, 0),
end=(1 / 3, 1),
sampling=probe.ctf.nyquist_sampling, # Equal to 0.36963 A
fractional=True,
potential=potential,
)fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12,6))
abtem.show_atoms(atoms, ax=ax1, plane="xy", title="Beam view")
abtem.show_atoms(atoms, ax=ax2, plane="yz", title="Side view", legend=True);
scan.add_to_plot(ax1);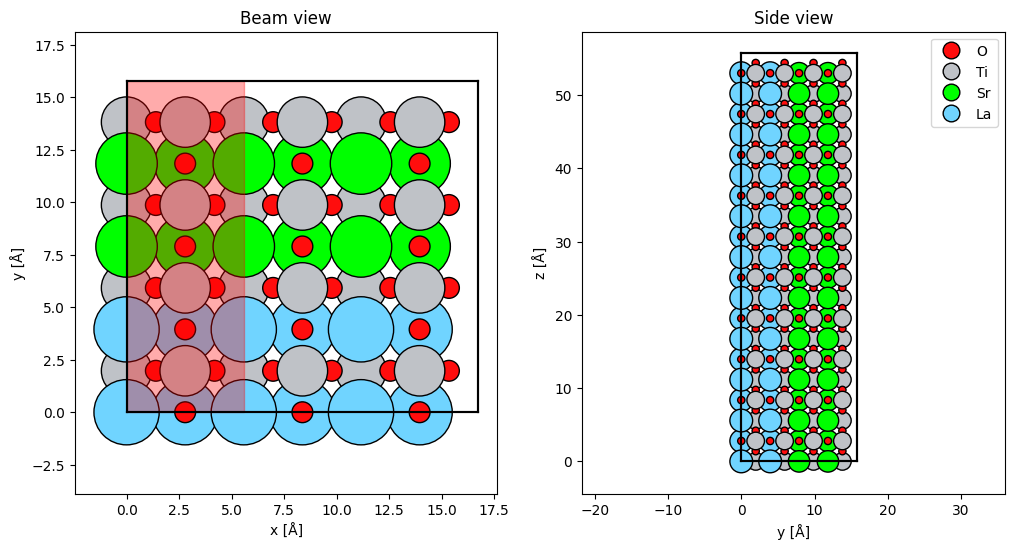
bright = abtem.AnnularDetector(inner=0, outer=20)
maadf = abtem.AnnularDetector(inner=40, outer=100)
haadf = abtem.AnnularDetector(inner=100, outer=180)
detectors = [bright, maadf, haadf]scanned_measurements = probe.scan(
scan=scan,
detectors=detectors,
potential=potential,
)
compute = False
if compute:
scanned_measurements.compute()
stacked_measurements = abtem.stack(scanned_measurements, ("BF", "MAADF", "HAADF"))
stacked_measurements.to_zarr("data/STO_LTO_STEM.zarr");
else:
stacked_measurements = abtem.from_zarr("data/STO_LTO_STEM.zarr").compute()
#stacked_measurements.show(explode=True);
#plt.savefig("../static/stem_images.png", dpi=600)[########################################] | 100% Completed | 105.71 ms
interpolated_measurements = stacked_measurements.interpolate(0.05)
#interpolated_measurements.show(explode=True);blurred_measurements = interpolated_measurements.gaussian_filter(0.35)
#blurred_measurements.show(explode=True);noisy_measurements = blurred_measurements.poisson_noise(dose_per_area=1e5, seed=100)
#noisy_measurements.show(explode=True, figsize=(12, 4));processed_measurements = abtem.stack([interpolated_measurements, blurred_measurements, noisy_measurements],
("Interpolated", "Blurred", "Noised"))# Images need to have the same number of pixels so we can stack them for easy visualization:
from scipy.ndimage import zoom
scalex = interpolated_measurements[0].shape[0]/stacked_measurements[0].array.shape[0]
scaley = interpolated_measurements[1].shape[1]/stacked_measurements[0].array.shape[1]
zoomed0 = zoom(stacked_measurements[0].array, (scalex, scaley), order=0)
zoomed1 = zoom(stacked_measurements[1].array, (scalex, scaley), order=0)
zoomed2 = zoom(stacked_measurements[2].array, (scalex, scaley), order=0)
from abtem import Images
image0 = Images(zoomed0, sampling = interpolated_measurements[0].sampling)
image1 = Images(zoomed1, sampling = interpolated_measurements[1].sampling)
image2 = Images(zoomed2, sampling = interpolated_measurements[2].sampling)
zoomed_measurements = abtem.stack([image0, image1, image2], ("BF", "MAADF", "HAADF"))all_measurements = abtem.stack([zoomed_measurements, interpolated_measurements, blurred_measurements, noisy_measurements],
("Nyquist-sampled", "Interpolated", "Blurred", "Noised"))
all_measurements.to_zarr("data/processed_STEM.zarr");[########################################] | 100% Completed | 101.45 ms
%matplotlib ipympl
abtem.config.set({"visualize.cmap": "viridis"})
abtem.config.set({"visualize.continuous_update": True})
abtem.config.set({"visualize.autoscale": True})
all_measurements = abtem.from_zarr("data/processed_STEM.zarr")viz = all_measurements.show(interact=True, explode=(1,), figsize=(5,5))[########################################] | 100% Completed | 106.16 ms
ImageGUI(children=(VBox(children=(SelectionSlider(options=('Nyquist-sampled', 'Interpolated', 'Blurred', 'Nois…
