CtrVAE
The Variational Autoencoder (VAE) is a type of deep generative model that can learn to encode high-dimensional data, such as images, into a low-dimensional latent space and then decode that latent representation back to the original data space. A VAE is particularly useful in imaging data, as it can capture meaningful features in a compressed form, making it easier to analyze patterns, generate new images, or explore variations in the data.
What Does a Simple VAE Do?
Encoder:
The encoder maps the input image into a latent space by compressing it into a lower-dimensional representation. Unlike a traditional autoencoder, which might produce a fixed vector, the VAE encoder outputs two components for each latent dimension: a mean and a log variance. These parameters define a Gaussian distribution over the latent space for each input.
Latent Space Sampling:
After the encoder produces a mean and variance, a sample is drawn from this Gaussian distribution, which allows the VAE to introduce some randomness or variability into the latent representation. The sampling process makes the VAE a generative model, enabling it to create new images by sampling different points in the latent space.
Decoder:
The sampled latent vector is then fed to the decoder, which reconstructs the image. The decoder tries to reproduce the original input as accurately as possible, allowing the VAE to learn a compressed, yet informative, representation of the input data.
Loss Function:
The VAE optimizes two components: Reconstruction Loss: Measures the similarity between the input image and the reconstructed image, encouraging the VAE to accurately capture image details. KL Divergence: Regularizes the latent space, ensuring the learned latent distributions are close to a standard Gaussian. This keeps the latent space smooth, meaning that similar points in the latent space correspond to similar reconstructed images.
from atomai import stat as atomstat
import atomai as aoi
import numpy as np
import pyroved as pv
import gdown
import torch
import random
tt = torch.tensor
torch.manual_seed(0)
# torch.cuda.manual_seed_all(0)
# torch.backends.cudnn.deterministic=True
np.random.seed(0)
random.seed(0)
import os
import wget
from sklearn.preprocessing import StandardScaler
import h5py
import matplotlib.pyplot as plt
from sklearn.mixture import GaussianMixture
from sklearn.decomposition import PCA
from skimage import feature
import skimage
from scipy.ndimage import zoom
from matplotlib.patches import Rectangle
import seaborn as sns
import ipywidgets as widgets
import ipywidgets
import pickle
from IPython.display import display, HTML
from ipywidgets import interact, Layout
from scipy.ndimage import zoom# ! gdown --fuzzy --id 1AHlk5xxXiuiTtYNr8fk0YQ8Uxjbf8bfT# Load the lists from the pickle file
images_data = "data/images_data.pkl"
with open(images_data, "rb") as f:
selected_images, ground_truth_px, ground_truth_py = pickle.load(f)
# Confirm successful loading by checking the lengths of the lists
print(len(selected_images), len(ground_truth_px), len(ground_truth_py))5 5 5
# min-max normalization:
def norm2d(img: np.ndarray) -> np.ndarray:
return (img - np.min(img)) / (np.max(img) - np.min(img))image = selected_images[0]
img = norm2d(image)def custom_extract_subimages(imgdata, coordinates, w_prime):
# Stage 1: Extract subimages with a fixed size (64x64)
large_window_size = (64, 64)
half_height_large = large_window_size[0] // 2
half_width_large = large_window_size[1] // 2
subimages_largest = []
coms_largest = []
for coord in coordinates:
cx = int(np.around(coord[0]))
cy = int(np.around(coord[1]))
top = max(cx - half_height_large, 0)
bottom = min(cx + half_height_large, imgdata.shape[0])
left = max(cy - half_width_large, 0)
right = min(cy + half_width_large, imgdata.shape[1])
subimage = imgdata[top:bottom, left:right]
if subimage.shape[0] == large_window_size[0] and subimage.shape[1] == large_window_size[1]:
subimages_largest.append(subimage)
coms_largest.append(coord)
# Stage 2: Use these centers to extract subimages of window size `w1`
half_height = w_prime[0] // 2
half_width = w_prime[1] // 2
subimages_target = []
coms_target = []
for coord in coms_largest:
cx = int(np.around(coord[0]))
cy = int(np.around(coord[1]))
top = max(cx - half_height, 0)
bottom = min(cx + half_height, imgdata.shape[0])
left = max(cy - half_width, 0)
right = min(cy + half_width, imgdata.shape[1])
subimage = imgdata[top:bottom, left:right]
if subimage.shape[0] == w_prime[0] and subimage.shape[1] == w_prime[1]:
subimages_target.append(subimage)
coms_target.append(coord)
return np.array(subimages_target), np.array(coms_target)def build_descriptor(window_size, min_sigma, max_sigma, threshold, overlap):
processed_img = img
all_atoms = skimage.feature.blob_log(processed_img, min_sigma, max_sigma, 30, threshold, overlap)
coordinates = all_atoms[:, : -1]
# Extract subimages
subimages_target, coms_target = custom_extract_subimages(processed_img, coordinates, window_size)
# Build descriptors
descriptors = [subimage.flatten() for subimage in subimages_target]
descriptors = np.array(descriptors)
return descriptors, coms_target, all_atoms, coordinates, subimages_targetNow we know the optimum hyperparameters
window_size = (40,40)
min_sigma = 1
max_sigma = 5
threshold = 0.025
overlap = 0.0
descriptors, coms_target, all_atoms, coordinates, subimages_target = build_descriptor(window_size, min_sigma, max_sigma, threshold, overlap)print(descriptors.shape)
print(coms_target.shape)
print(all_atoms.shape)
print(coordinates.shape)
print(subimages_target.shape)(10917, 1600)
(10917, 2)
(11813, 3)
(11813, 2)
(10917, 40, 40)
However, our analysis above had a signnificant limitation. We were looking for latent representation common for A- and B-site cations, even though these are obviously different. Now, we are going to explore the conditional VAE, that consider cation type as known variable. Let’s see how it works!
# Define the Fit_GMM_param function without PCA, including covariance type
def Fit_GMM(descriptors, components, covariance_type):
# First pass of GMM to estimate initial parameters
# Flatten each subimage into a 1D vector
flattened_descriptors = descriptors.reshape(descriptors.shape[0], -1)
# Remove subimages with NaN values
mask = ~np.isnan(flattened_descriptors).any(axis=1)
valid_subimages = flattened_descriptors[mask]
preliminary_gmm = GaussianMixture(n_components=components, covariance_type=covariance_type, random_state=42)
preliminary_gmm.fit(valid_subimages)
initial_means = preliminary_gmm.means_
initial_weights = preliminary_gmm.weights_
# Initialize and fit the GMM using the parameters from the preliminary GMM
gmm = GaussianMixture(n_components=components,
means_init=initial_means,
weights_init=initial_weights,
covariance_type=covariance_type,
random_state=42)
gmm.fit(valid_subimages)
# Map the labels back to the original data, including NaN-handling
labels = gmm.predict(valid_subimages)
full_labels = np.full(valid_subimages.shape[0], -1)
full_labels[mask] = labels
return labels, valid_subimagesw = (40,40)
min_sigma = 1
max_sigma = 5
threshold = 0.025
overlap = 0.0
descriptors_c, coms_target_c, all_atoms_c, coordinates_c, subimages_target_c = build_descriptor(w,
min_sigma,
max_sigma,
threshold,
overlap)
print(descriptors_c.shape)
print(subimages_target_c.shape)
print(coms_target_c.shape)
print(all_atoms_c.shape)
print(coordinates_c.shape)
labels_c, valid_subimages = Fit_GMM(descriptors_c, 2, "full")
print(labels_c.shape)(10917, 1600)
(10917, 40, 40)
(10917, 2)
(11813, 3)
(11813, 2)
(10917,)
plt.figure(figsize=(6, 6))
plt.imshow(image, cmap='gray')
plt.scatter(coms_target_c[:, 1], coms_target_c[:, 0], c=labels_c, s=40, cmap='cool', marker= "o")
plt.xlim([400,700])
plt.ylim([400,700])
plt.axis('off')
plt.show()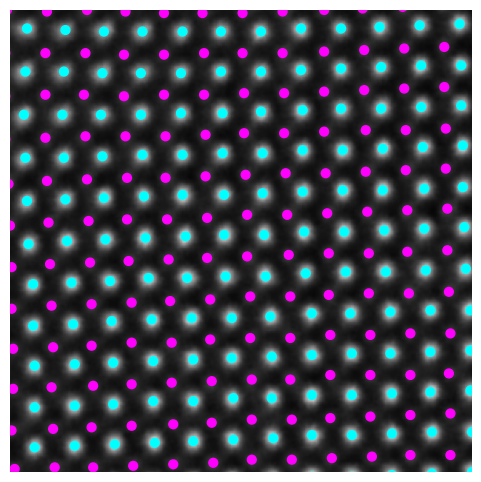
subimages_target_reshaped = np.expand_dims(subimages_target, axis=-1)
train_data = torch.tensor(subimages_target_reshaped[:,:,:,0]).float()
lab_train = pv.utils.to_onehot(tt(labels_c).to(torch.int64), 2)
train_loader = pv.utils.init_dataloader(train_data.unsqueeze(1), lab_train, batch_size=48)Now, running the VAE in PyroVEd. Simple VAE will find the best representation of our data as two components for latent vecotr (l1,l2). Of course, we can explore other dimensinalities of latent space!
# in_dim = (window_size[0],window_size[1])
# # Initialize vanilla VAE
# Crvae = pv.models.iVAE(in_dim, latent_dim=2, c_dim = 2, # Number of latent conditions
# hidden_dim_e = [512,512], # corresponds to the number of neurons in the hidden layers of the encoder
# hidden_dim_d = [512,512],# corresponds to the number of neurons in the hidden layers of the decoder
# # corresponds to the number of neurons in the hidden layers of the decoder
# invariances=["r"], seed=0)
# # Initialize SVI trainer
# trainer = pv.trainers.SVItrainer(Crvae)
# # Train for n epochs:
# for e in range(10):
# trainer.step(train_loader)
# trainer.print_statistics()
# Crvae.save_weights('crvae_model')
# print("Model saved successfully.")# ! gdown --fuzzy --id 1xCTjijGlBADpIFmeRBP-qH6FQMspGGdUin_dim = (window_size[0],window_size[1])
# Reinitialize the model before loading weights
Ctrvae_model = pv.models.iVAE(in_dim, latent_dim=2, c_dim = 2, # Number of latent dimensions other than the invariancies
hidden_dim_e = [512, 512],
hidden_dim_d = [512, 512], # corresponds to the number of neurons in the hidden layers of the decoder
invariances=["r", "t"], seed=0)
# Load the saved model weights
Ctrvae_model.load_weights('data/ctrvae_model.pt')
print("Model loaded successfully.")Model loaded successfully.
Varitional Auto Encoder manifold representation
Ctrvae_z_mean, Ctrvae_z_sd = Ctrvae_model.encode(train_data.unsqueeze(-1), lab_train)
print('no. of defects', Ctrvae_z_mean.shape)
z1 = Ctrvae_z_mean[:, -2]
z2 = Ctrvae_z_mean[:, -1]
ang = Ctrvae_z_mean[:, 0]
tx = Ctrvae_z_mean[:, -4]
ty = Ctrvae_z_mean[:, -3]no. of defects torch.Size([10917, 5])
def generate_latent_manifold_C(n=10, decoder=None, target_size=(28, 28), condition=None):
# Define grid bounds across latent space
grid_x = np.linspace(min(z1), max(z1), n)
grid_y = np.linspace(min(z2), max(z2), n)
# Dynamically infer output shape
latent_sample = torch.tensor([[grid_x[0], grid_y[0]]], dtype=torch.float32)
if condition is not None:
latent_sample = torch.cat((latent_sample, condition), dim=1)
with torch.no_grad():
X_decoded = decoder(latent_sample)
# Check output dimensions
if len(X_decoded.shape) == 1:
X_decoded = X_decoded.reshape(1, -1) # Flattened output
output_size = X_decoded.shape[-1] if len(X_decoded.shape) == 2 else X_decoded.shape[-2:]
# Initialize the manifold
height, width = target_size
manifold = np.zeros((height * n, width * n))
# Generate the manifold grid
for i, yi in enumerate(grid_x):
for j, xi in enumerate(grid_y):
latent_sample = torch.tensor([[xi, yi]], dtype=torch.float32)
if condition is not None:
latent_sample = torch.cat((latent_sample, condition), dim=1)
with torch.no_grad():
X_decoded = decoder(latent_sample)
if len(X_decoded.shape) == 1:
X_decoded = X_decoded.reshape(1, -1) # Handle flattened output
resized_image = zoom(X_decoded.numpy().reshape(output_size),
zoom=(height / output_size[0], width / output_size[1]))
manifold[i * height: (i + 1) * height, j * width: (j + 1) * width] = resized_image
return manifoldThe latent representation of the system is visualized as a grid over the two latent variables and . Each grid cell corresponds to a unique combination of values for and , which are decoded to produce corresponding reconstructions in the data space. The smooth and structured transition across the grid indicates that the model has learned a meaningful and continuous mapping between the latent variables and the data space. Variations in the grid reflect changes in the underlying physical structure, such as column type, domain orientation, or material properties.
display(HTML("""
<style>
.widget-label { font-size: 16px; font-weight: bold; }
select { font-size: 16px; font-weight: bold; }
</style>
"""))
# Define dropdown styling
dropdown_style = {'description_width': 'initial'}
dropdown_layout = Layout(width='250px')
# **Dropdown options**
options_A = ["Manifold 1", "Manifold 2"] # Panel A options
options_B = ["z1", "z2", "angle", "tx", "ty"] # Panel B (X & Y variables)
# **Generate Latent Manifold with Conditioning**
def generate_latent_manifold_C(n=10, decoder=None, target_size=(40, 40), condition=None):
grid_x = np.linspace(min(z1), max(z1), n)
grid_y = np.linspace(min(z2), max(z2), n)
height, width = target_size
manifold = np.zeros((height * n, width * n))
for i, yi in enumerate(grid_x):
for j, xi in enumerate(grid_y):
latent_sample = torch.tensor([[xi, yi]], dtype=torch.float32)
if condition is not None:
latent_sample = torch.cat((latent_sample, condition), dim=1)
with torch.no_grad():
X_decoded = decoder(latent_sample)
resized_image = X_decoded.numpy().reshape(target_size)
manifold[i * height: (i + 1) * height, j * width: (j + 1) * width] = resized_image
return manifold
# **Interactive Plot Function**
def interactive_CVAE(panel_A, variable_x, variable_y):
"""Creates an interactive plot with selectable Manifold (A) and scatter/KDE (B)."""
fig, axes = plt.subplots(1, 2, figsize=(12, 6))
# **Panel A (Left) - Selectable Manifold**
condition_0 = torch.nn.functional.one_hot(torch.tensor([0]), num_classes=2).float()
condition_1 = torch.nn.functional.one_hot(torch.tensor([1]), num_classes=2).float()
if panel_A == "Manifold 1":
manifold = generate_latent_manifold_C(n=10, decoder=Ctrvae_model.decode, target_size=(40, 40), condition=condition_0)
elif panel_A == "Manifold 2":
manifold = generate_latent_manifold_C(n=10, decoder=Ctrvae_model.decode, target_size=(40, 40), condition=condition_1)
axes[0].imshow(manifold, cmap="gnuplot2", origin="upper", aspect="auto")
axes[0].set_xlabel(r"$z_1$", fontsize=16, fontweight="bold")
axes[0].set_ylabel(r"$z_2$", fontsize=16, fontweight="bold")
axes[0].set_xticks([]), axes[0].set_yticks([])
axes[0].text(-0.05, 1, 'a)', transform=axes[0].transAxes, fontsize=16, fontweight='bold', va='top', ha='right')
# **Panel B (Right) - Interactive Scatter/KDE Plot**
variable_map = {
"z1": (z1, r"$z_1$", "plasma", "cyan"),
"z2": (z2, r"$z_2$", "plasma", "magenta"),
"angle": (ang, r"$\theta$", "plasma", "blue"),
"tx": (tx, r"$t_x$", "inferno", "green"),
"ty": (ty, r"$t_y$", "inferno", "orange")
}
var_x, label_x, cmap_x, color_x = variable_map[variable_x]
var_y, label_y, cmap_y, color_y = variable_map[variable_y]
sns.scatterplot(x=var_x, y=var_y, ax=axes[1], hue=labels_c, alpha=0.4, edgecolor="k", s=10)
try:
sns.kdeplot(x=var_x, y=var_y, ax=axes[1], cmap="Oranges", levels=50, thresh=0.05, alpha=0.4, fill=False, warn_singular=False)
except ValueError:
pass
axes[1].set_xlabel(label_x, fontsize=16, fontweight="bold")
axes[1].set_ylabel(label_y, fontsize=16, fontweight="bold")
axes[1].text(-0.05, 1, 'b)', transform=axes[1].transAxes, fontsize=16, fontweight='bold', va='top', ha='right')
# Adjust layout for better spacing
plt.tight_layout()
plt.show()<IPython.core.display.HTML object># **Create interactive dropdown widgets**
interact(interactive_CVAE,
panel_A=widgets.Dropdown(options=options_A, description="Panel A", layout=Layout(width='250px')),
variable_x=widgets.Dropdown(options=options_B, description="X-Axis Variable", layout=Layout(width='250px')),
variable_y=widgets.Dropdown(options=options_B, description="Y-Axis Variable", layout=Layout(width='250px'))
);interactive(children=(Dropdown(description='Panel A', layout=Layout(width='250px'), options=('Manifold 1', 'Ma…# Apply styling for dropdowns
display(HTML("""
<style>
.widget-label { font-size: 16px; font-weight: bold; }
select { font-size: 16px; font-weight: bold; }
</style>
"""))
# **Dropdown Styling**
dropdown_style = {'description_width': 'initial'}
dropdown_layout = Layout(width='250px')
# **Available Options**
options = ["z1", "z2", "tx", "ty", "Ground Truth Px", "Ground Truth Py", "Angle"]
# **Define Variables**
Px = ground_truth_px[0]
Py = ground_truth_py[0]
# **Define a dictionary for mapping options to data and plot type**
plot_data = {
"z1": {"data": z1, "type": "scatter"},
"z2": {"data": z2, "type": "scatter"},
"tx": {"data": tx, "type": "scatter"},
"ty": {"data": ty, "type": "scatter"},
"Ground Truth Px": {"data": Px, "type": "image"},
"Ground Truth Py": {"data": Py, "type": "image"},
"Angle": {"data": ang, "type": "scatter"}
}
def plot_variable(ax, variable, subplot_label):
"""Plots the selected variable in the given axis."""
data = plot_data[variable]["data"]
plot_type = plot_data[variable]["type"]
if plot_type == "scatter":
ax.scatter(coms_target[:, 1], coms_target[:, 0], c=data, s=14, cmap='jet', marker="o")
elif plot_type == "image":
ax.imshow(data, cmap='jet', origin='lower')
ax.axis("off")
ax.text(-0.05, 1, subplot_label, transform=ax.transAxes, fontsize=16, fontweight='bold', va='top', ha='right')
def plot_two_variables(variable1, variable2):
"""Creates a 1-row, 2-column figure and plots two selected variables."""
fig, axes = plt.subplots(1, 2, figsize=(12, 6))
plot_variable(axes[0], variable1, 'a)')
plot_variable(axes[1], variable2, 'b)')
plt.tight_layout()
plt.show()<IPython.core.display.HTML object># **Create interactive dropdown widgets**
interact(plot_two_variables,
variable1=widgets.Dropdown(options=options, description="Variable 1", layout=dropdown_layout),
variable2=widgets.Dropdown(options=options, description="Variable 2", layout=dropdown_layout)
);interactive(children=(Dropdown(description='Variable 1', layout=Layout(width='250px'), options=('z1', 'z2', 't…
