Deep Learning Applications in Microscopy: Segmentation and Tracking
Discussion
Segmentation and Tracking¶
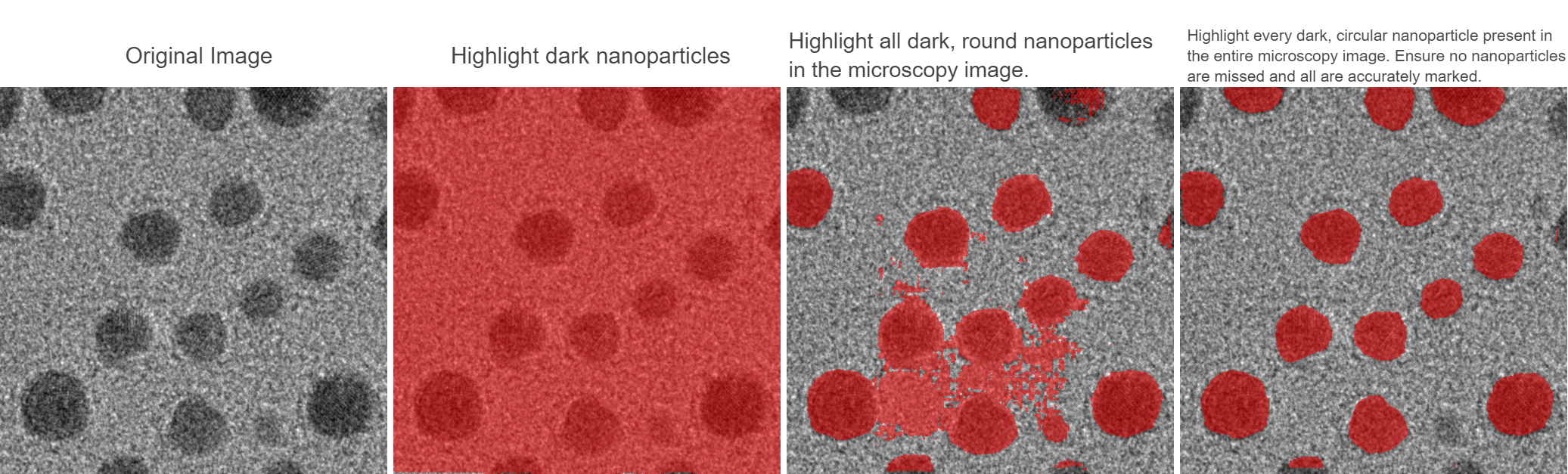
The models released for computer vision are often based on realistic images, which are often ineffective when applied directly to microscope data for materials research. This paper takes microscope datasets and trains or fine-tunes them on different models. Deep learning shows the superior performance of the models in microscope image segmentation. All four segmentation models perform well on both the training and test sets, especially the EfficientSAM model, which shows the highest stability and accuracy on all evaluation metrics, indicating its strong generalization ability and robustness in the segmentation task. EfficientSAM demonstrates better segmentation performance in the application scenario Figure 3.3, and the edges are closer to the ground truth, according to the results in Table 3.1, EfficientSAM-tiny does have an obvious advantage in terms of accuracy. Although the other models have their own characteristics in terms of throughput, number of parameters and memory usage, EfficientSAM-tiny outperforms the other models in terms of accuracy, with an IoU of 0.99672 and a Dice Coefficient of 0.99836. At the same time, the performance of EfficientSAM model is better than the other models in terms of loss in both the training and the testing curves, showing optimal segmentation performance and generalization ability. These results suggest that the EfficientSAM model may be a potentially superior choice for handling segmentation tasks with greater efficiency and effectiveness. This provides an important reference for the selection and optimization of the tracking model.
For the autoencoder architecture, the encoder part of the network drastically reduces the resolution of the feature maps through the pooling layer, which is not conducive to generating accurate segmentation masks. Skip-connection in the Swin-UNet can introduce high-resolution features from the shallow convolutional layers, which contain rich low-level information to help generate better segmentation masks. However, the edges of the mask after Swin-UNet segmentation are still not clean enough in Figure 3.3. The algorithms in the tracking part are able to maintain high performance when dealing with the kinematic complexities such as particle growth, and motion during sintering, demonstrating the dynamics of the particles. This indicates the tracking method of this paper has significant application potential. In particular, the outstanding EfficientSAM and the efficiency of the DeAOT model demonstrate the potential of deep learning techniques in microscopy image/video analysis.
Future Directions¶
In Figure 3.1, the training loss of the YOLO model decreases initially, but the test loss picks up after reaching its lowest point in the 580 epoch and shows a U-shaped curve. This indicates that the model is overfitting. This may be due to the fact that a model of this size is not able to capture the visual information in the image well. However, this does not mean that the model is not useful. The advantage of YOLO is the speed of inference. In future research, YOLO can be used to perform initial segmentation at high speed to obtain positional information, and then SAM could be used to re-segment the critical parts. This will increase the speed of inference while still maintaining high accuracy. In addition, in higher pixel resolution microscope videos, it may be necessary to cut the image into small regions for tracking, which may significantly slow down the inference speed of the model. In this case, YOLO can be used to perform overall fast target detection on the scaled resolution image, and then the high pixel resolution target region can be cropped, based on the detection result. Thereafter, SAM can be used to obtain more accurate segmentation.
In the future, there are potential applications of large language modeling (LLM) in microscopy image analysis. For example, the “Large Language Instructed Segmentation Assistant” (LISA) model is fine-tuned with a multimodal large language model to reason and generate “tokens” with approximate location information and combined with the SAM model for accurate segmentation Lai et al., 2024. However, the LISA model still has some limitations when processing microscopy images. Although the segmentation level can be improved by optimizing the prompt, as shown in top of Figure 4.1, although most of the nanoparticles are correctly labeled, there are still some particles that are missed or incorrectly labeled. The main reason for this is that the training data is mainly from real-world scenarios rather than specialized microscope images. Perhaps incorporating microscope images into the training data could help improve the model’s performance in microscope images. In another study Zhang et al., 2023 on sam-guided multimodal LLM, the SAM visual coder was simultaneously introduced into a multimodal large language model. By adding a SAM visual encoder (for detail information) to the CLIP visual encoder (for global information) more detailed characterization can be achieved. Using multiple visual encoders with different functions can significantly improve the accuracy and detail of the generated image descriptions Zhang et al., 2023. Visual encoders with different functions are similar to the compound eyes of an insect and can provide information with different details: the CLIP visual encoder provides the overall category and information of the image, while the SAM visual encoder provides information about the edges and shapes of the objects. In the future, by combining the multimodal LLM model with the SAM model, it is expected to further enhance the ability to analyze and reason about microscope videos.
- Lai, X., Tian, Z., Chen, Y., Li, Y., Yuan, Y., Liu, S., & Jia, J. (2024). LISA: Reasoning Segmentation via Large Language Model. 10.48550/arXiv.2308.00692
- Zhang, Z., Wang, B., Liang, W., Li, Y., Guo, X., Wang, G., Li, S., & Wang, G. (2023). Sam-Guided Enhanced Fine-Grained Encoding with Mixed Semantic Learning for Medical Image Captioning. 10.48550/arXiv.2311.01004