Contents
Multi-Modal Tutorial Co3O4-Mn3O4
Guided Computation of Fused Multi-Modal Electron Microscopy¶
This tutorial is almost identical to the previous, but now we use a new Co3O4-Mn3O4 dataset which utilizes EELS instead of X-EDS. We also read from a .h5 file in a similar fashion to how one would read from a .dm3, .dm4, or .emd file format. The parameters for convergence have also changed slightly, highlighting how one set of weights may not work across datasets, hence assessing cost function convergence and regularization weighting is key. Just like the previous dataset, dramatic improvement in image quality is observed within just a few minutes of parameter tuning as seen in Figure 4.1
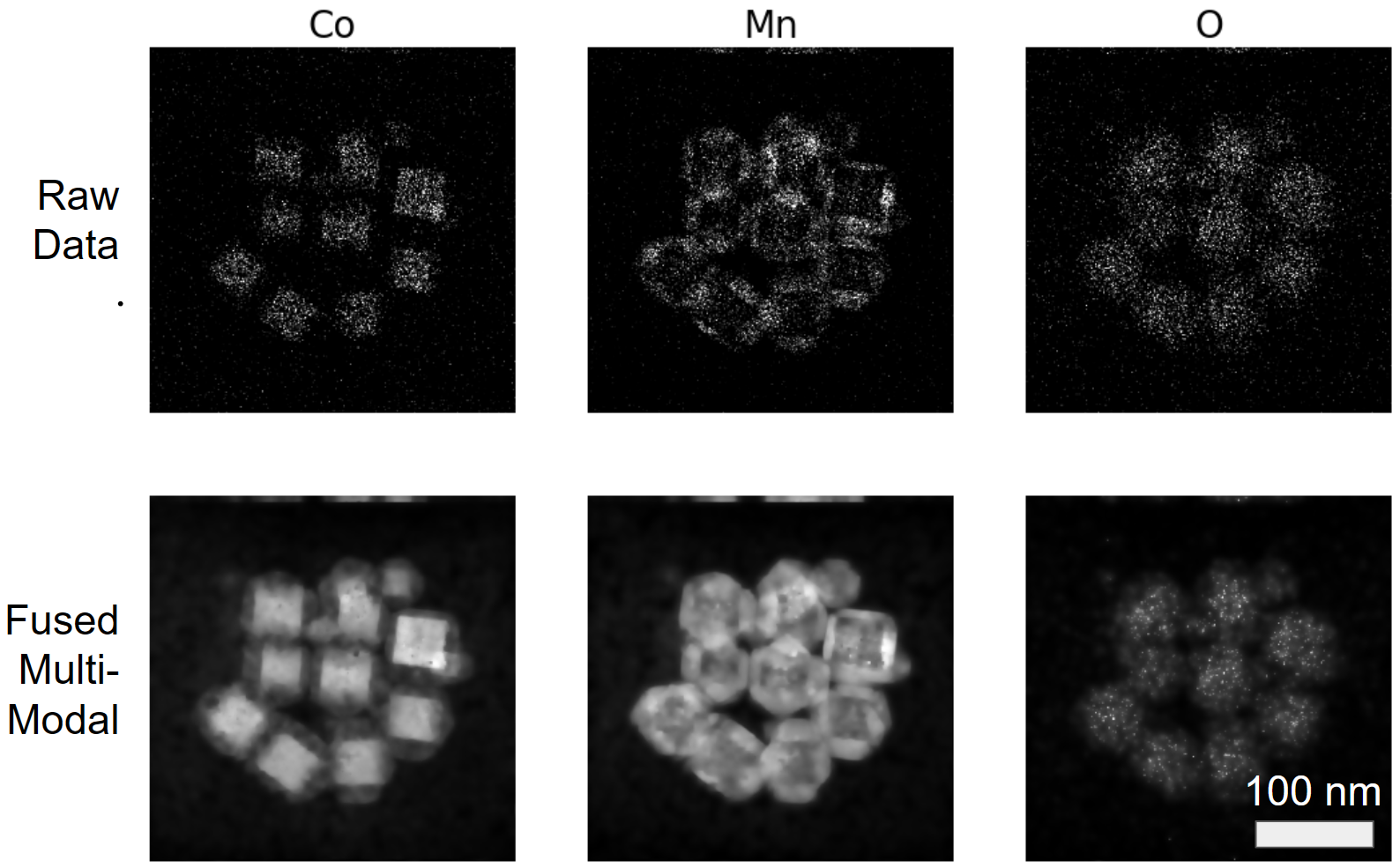
Figure 4.1:Comparison of raw input vs fused multi-modal Co3O4-Mn3O4 HAADF elastic and X-EDS inelastic images
import data.fusion_utils as utils
from data.widget_helpers import return_reconstruction_plots
from scipy.sparse import spdiags
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
import numpy as np
import h5py
import ipywidgets as widgets
from IPython.display import displaydata = 'data/Co3O4_Mn3O4.h5'
# Define element names and their atomic weights
elem_names=['Co', 'Mn', 'O']
elem_weights=[27,25,8]
# Parse elastic HAADF data and inelastic chemical maps based on element index from line above
with h5py.File(data, 'r') as h5_file:
HAADF = np.array(h5_file['HAADF'][:])
xx = np.array([],dtype=np.float32)
for ee in elem_names:
# Read chemical maps
with h5py.File(data, 'r') as h5_file:
chemMap = np.array(h5_file[ee][:])
# Check if chemMap has the same dimensions as HAADF
if chemMap.shape != HAADF.shape:
raise ValueError(f"The dimensions of {ee} chemical map do not match HAADF dimensions.")
# Set Noise Floor to Zero and Normalize Chemical Maps
chemMap -= np.min(chemMap); chemMap /= np.max(chemMap)
# Concatenate Chemical Map to Variable of Interest
xx = np.concatenate([xx,chemMap.flatten()])# Make Copy of Raw Measurements for Poisson Maximum Likelihood Term
xx0 = xx.copy()
# Incoherent linear imaging for elastic scattering scales with atomic number Z raised to γ ∈ [1.4, 2]
gamma = 1.6
# Image Dimensions
(nx, ny) = chemMap.shape; nPix = nx * ny
nz = len(elem_names)
# C++ TV Min Regularizers
reg = utils.tvlib(nx,ny)
# Data Subtraction and Normalization
HAADF -= np.min(HAADF); HAADF /= np.max(HAADF)
HAADF=HAADF.flatten()
# Create Summation Matrix
A = utils.create_weighted_measurement_matrix(nx,ny,nz,elem_weights,gamma,1)fig, ax = plt.subplots(1, nz + 1, figsize=(15, 8)) # Updated to accommodate an additional subplot for HAADF
ax = ax.flatten()
for ii in range(nz):
ax[ii].imshow(xx0[ii*(nx*ny):(ii+1)*(nx*ny)].reshape(nx, ny), cmap='gray', vmax=0.3)
ax[ii].set_title(elem_names[ii])
ax[ii].axis('off')
ax[nz].imshow(HAADF.reshape(nx, ny), cmap='gray')
ax[nz].set_title('HAADF')
ax[nz].axis('off')
fig.tight_layout()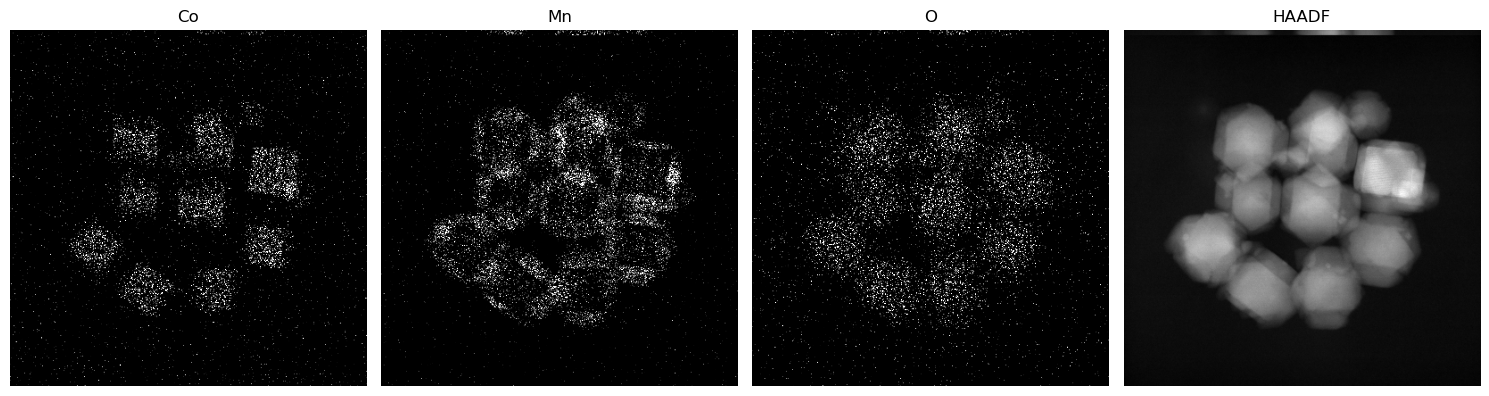
# Convergence Parameters
lambdaHAADF = 1/nz # Do not modify this
lambdaChem = 0.006
nIter = 30 # Typically 10-15 will suffice
lambdaTV = 0.022; #Typically between 0.001 and 1
bkg = 1e-2
# FGP TV Parameters
regularize = True; nIter_TV = 6; # xx represents the flattened 1D elastic maps we are trying to improve via the cost function
xx = xx0.copy()
# Background noise subtraction for improved convergence
xx = np.where((xx < .2), 0, xx)
# Auxiliary Functions for measuring the cost functions
lsqFun = lambda inData : 0.5 * np.linalg.norm(A.dot(inData**gamma) - HAADF) **2
poissonFun = lambda inData : np.sum(xx0 * np.log(inData + 1e-8) - inData)
# Main Loop
# Initialize the three cost functions components
costHAADF = np.zeros(nIter,dtype=np.float32); costChem = np.zeros(nIter, dtype=np.float32); costTV = np.zeros(nIter, dtype=np.float32);
for kk in tqdm(range(nIter)):
# Solve for the first two optimization functions $\Psi_1$ and $\Psi_2$
xx -= gamma * spdiags(xx**(gamma - 1), [0], nz*nx*ny, nz*nx*ny) * lambdaHAADF * A.transpose() * (A.dot(xx**gamma) - HAADF) + lambdaChem * (1 - xx0 / (xx + bkg))
# Enforce positivity constraint
xx[xx<0] = 0
# FGP Regularization if turned on
if regularize:
for zz in range(nz):
xx[zz*nPix:(zz+1)*nPix] = reg.fgp_tv( xx[zz*nPix:(zz+1)*nPix].reshape(nx,ny), lambdaTV, nIter_TV).flatten()
# Measure TV Cost Function
costTV[kk] += reg.tv( xx[zz*nPix:(zz+1)*nPix].reshape(nx,ny) )
# Measure $\Psi_1$ and $\Psi_2$ Cost Functions
costHAADF[kk] = lsqFun(xx); costChem[kk] = poissonFun(xx) 0%| | 0/30 [00:00<?, ?it/s]# Display Cost Functions and Descent Parameters
utils.plot_convergence(costHAADF, lambdaHAADF, costChem, lambdaChem, costTV, lambdaTV)
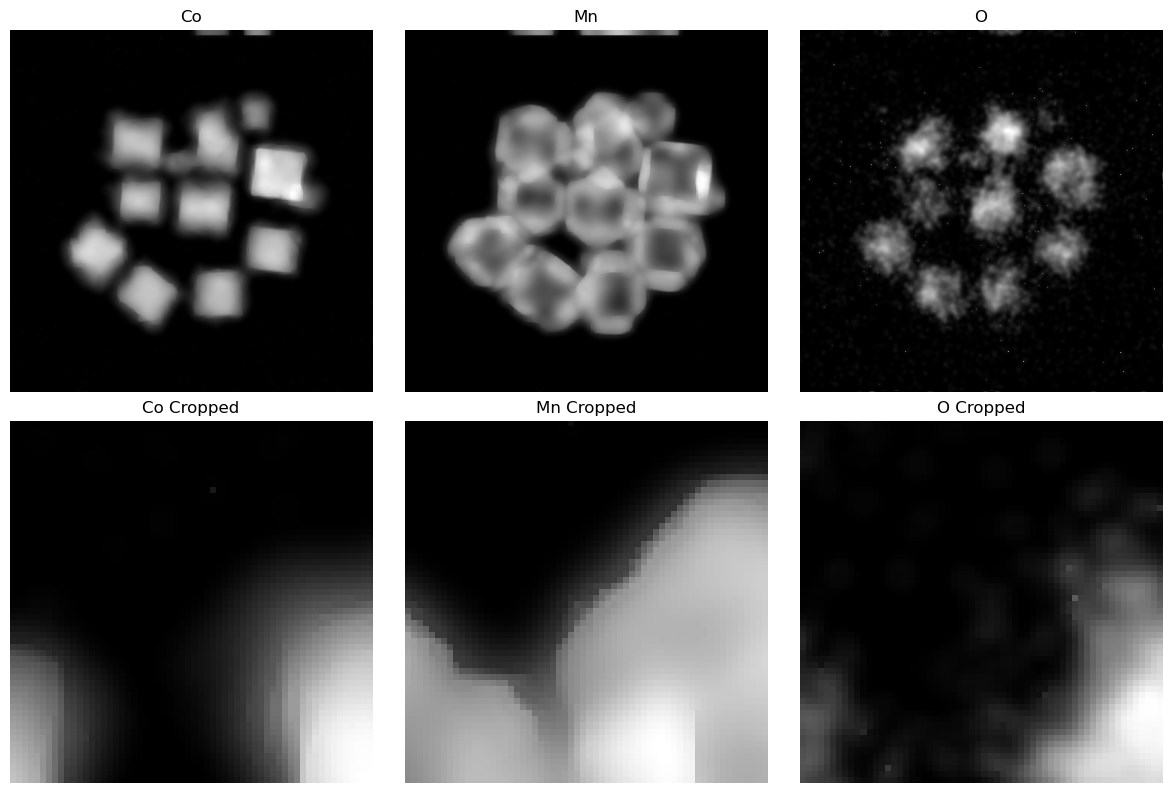
# Show Reconstructed Signal
fig, ax = plt.subplots(2, len(elem_names), figsize=(12, 8))
ax = ax.flatten()
for ii in range(len(elem_names)):
ax[ii].imshow(xx[ii*(nx*ny):(ii+1)*(nx*ny)].reshape(nx, ny), cmap='gray')
ax[ii].set_title(elem_names[ii])
ax[ii].axis('off')
ax[ii + len(elem_names)].imshow(xx[ii*(nx*ny):(ii+1)*(nx*ny)].reshape(nx, ny)[60:120, 160:220], cmap='gray')
ax[ii + len(elem_names)].set_title(elem_names[ii] + ' Cropped')
ax[ii + len(elem_names)].axis('off')
fig.tight_layout()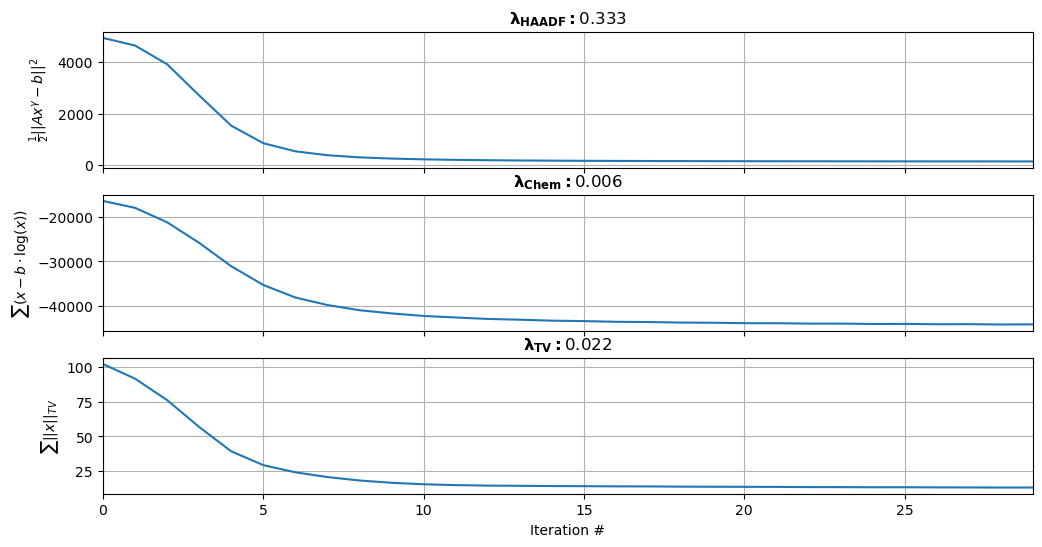
# Widgets for the parameters
kwargs = {
'style':{'description_width': 'initial'},
'layout':widgets.Layout(width='400px'),
'continuous_update': False,
'readout_format':'.3f'
}
lambdaChem_slider = widgets.FloatSlider(value=lambdaChem, min=0.001, max=1, step=0.001, description='lambdaChem',**kwargs)
lambdaTV_slider = widgets.FloatSlider(value=lambdaTV, min=0.001, max=1, step=0.001, description='lambdaTV',**kwargs)
nIter_slider = widgets.IntSlider(value=nIter, min=10, max=50, step=1, description='# Cost Function Iterations',**kwargs)
nIter_TV_slider = widgets.IntSlider(value=nIter_TV, min=1, max=10, step=1, description=' # TV Iterations',**kwargs)
def widget_wrapper(lambdaChem,lambdaTV,nIter,nIter_TV):
return_reconstruction_plots(
xx0,
HAADF,
A,
bkg,
(nx,ny,nz),
elem_names,
(60,120,160,220),
lambdaChem,
lambdaTV,
nIter,
nIter_TV,
subtract_bkg = 0.2
)
widgets.interact(widget_wrapper, lambdaChem=lambdaChem_slider, lambdaTV=lambdaTV_slider, nIter=nIter_slider, nIter_TV=nIter_TV_slider);interactive(children=(FloatSlider(value=0.006, continuous_update=False, description='lambdaChem', layout=Layou…#save_folder_name='test'
#utils.save_data(save_folder_name, xx0, xx, HAADF, A.dot(xx**gamma), elem_names, nx, ny, costHAADF, costChem, costTV, lambdaHAADF, lambdaChem, lambdaTV, gamma)
