Multi-Modal Tutorial DyScO3
Guided Computation of Fused Multi-Modal Electron Microscopy¶
This tutorial describes how you can fuse your EELS/X-EDS maps with HAADF or similar elastic imaging modalities to improve chemical resolution. This is Tutorial 1 of 2 where we look at an atomic resolution HAADF and X-EDS dataset of DyScO3. The multi-modal data fusion workflow relies on Python, and requires minimal user input with <10 tunable lines. Both here and in the Mathematical Overview section we outline best practices for these adjustments. Within a few minutes, datasets such as the one in this tutorial can be transformed into resolution enhanced chemical maps.
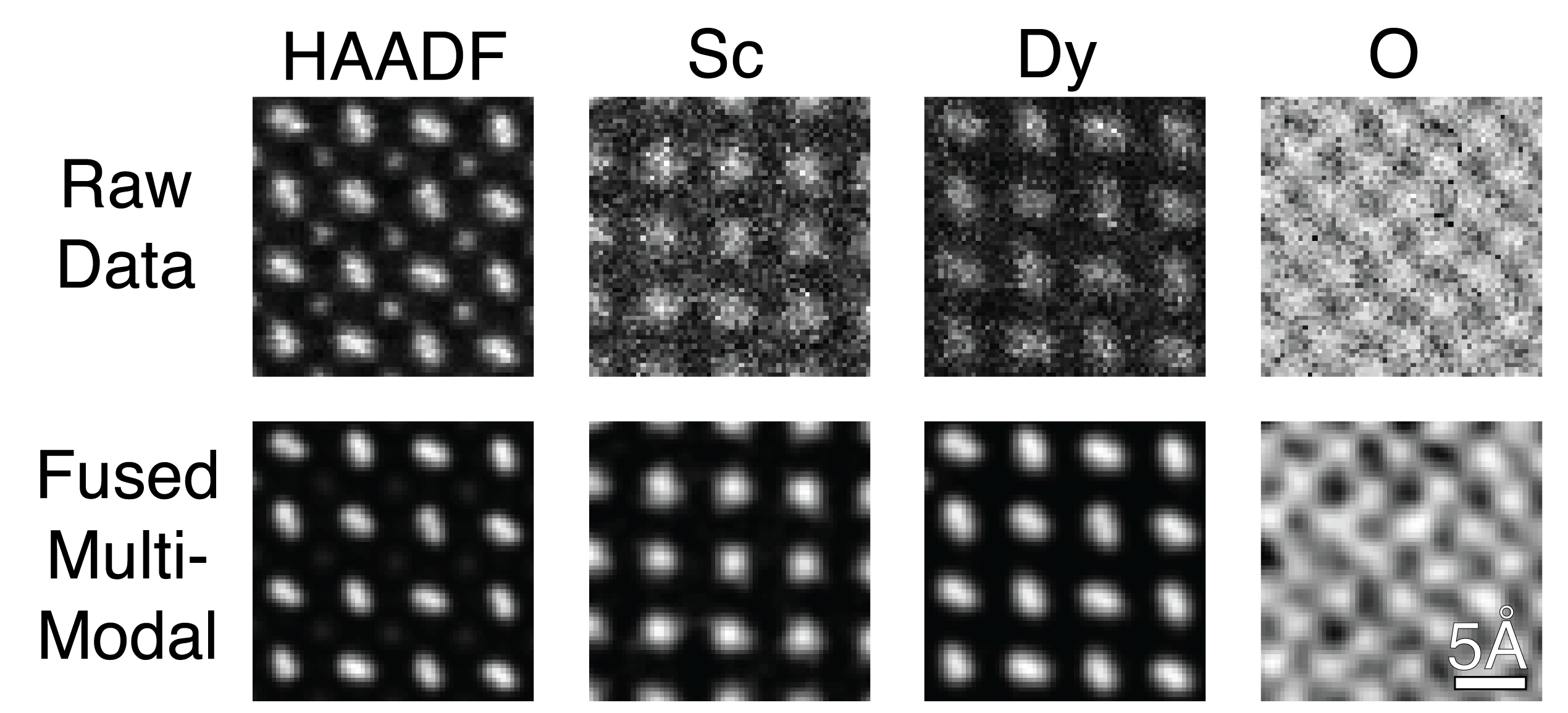
Figure 3.1:Comparison of raw input vs fused multi-modal DyScO3 HAADF elastic and X-EDS inelastic images
import data.fusion_utils as utils
from data.widget_helpers import return_reconstruction_plots
from scipy.sparse import spdiags
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
import numpy as np
import ipywidgets as widgets
from IPython.display import displaydata = np.load('data/PTO_Trilayer_dataset.npz')
# Define element names and their atomic weights
elem_names=['Sc', 'Dy', 'O']
elem_weights=[21,66,8]
# Parse elastic HAADF data and inelastic chemical maps based on element index from line above
HAADF = data['HAADF']
xx = np.array([],dtype=np.float32)
for ee in elem_names:
# Read Chemical Map for Element "ee"
chemMap = data[ee]
# Check if chemMap has the same dimensions as HAADF
if chemMap.shape != HAADF.shape:
raise ValueError(f"The dimensions of {ee} chemical map do not match HAADF dimensions.")
# Set Noise Floor to Zero and Normalize Chemical Maps
chemMap -= np.min(chemMap); chemMap /= np.max(chemMap)
# Concatenate Chemical Map to Variable of Interest
xx = np.concatenate([xx,chemMap.flatten()])# Make Copy of Raw Measurements for Poisson Maximum Likelihood Term
xx0 = xx.copy()
# Incoherent linear imaging for elastic scattering scales with atomic number Z raised to γ ∈ [1.4, 2]
gamma = 1.6
# Image Dimensions
(nx, ny) = chemMap.shape; nPix = nx * ny
nz = len(elem_names)
# C++ TV Min Regularizers
reg = utils.tvlib(nx,ny)
# Data Subtraction and Normalization
HAADF -= np.min(HAADF); HAADF /= np.max(HAADF)
HAADF=HAADF.flatten()
# Create Summation Matrix
A = utils.create_weighted_measurement_matrix(nx,ny,nz,elem_weights,gamma,1)fig, ax = plt.subplots(2,len(elem_names)+1,figsize=(12,6.5))
ax = ax.flatten()
ax[0].imshow(HAADF.reshape(nx,ny),cmap='gray'); ax[0].set_title('HAADF'); ax[0].axis('off')
ax[1+len(elem_names)].imshow(HAADF.reshape(nx,ny)[70:130,25:85],cmap='gray'); ax[1+len(elem_names)].set_title('HAADF Cropped'); ax[1+len(elem_names)].axis('off')
for ii in range(len(elem_names)):
ax[ii+1].imshow(xx0[ii*(nx*ny):(ii+1)*(nx*ny)].reshape(nx,ny),cmap='gray')
ax[ii+2+len(elem_names)].imshow(xx0[ii*(nx*ny):(ii+1)*(nx*ny)].reshape(nx,ny)[70:130,25:85],cmap='gray')
ax[ii+1].set_title(elem_names[ii])
ax[ii+1].axis('off')
ax[ii+2+len(elem_names)].set_title(elem_names[ii]+' Cropped')
ax[ii+2+len(elem_names)].axis('off')
fig.tight_layout()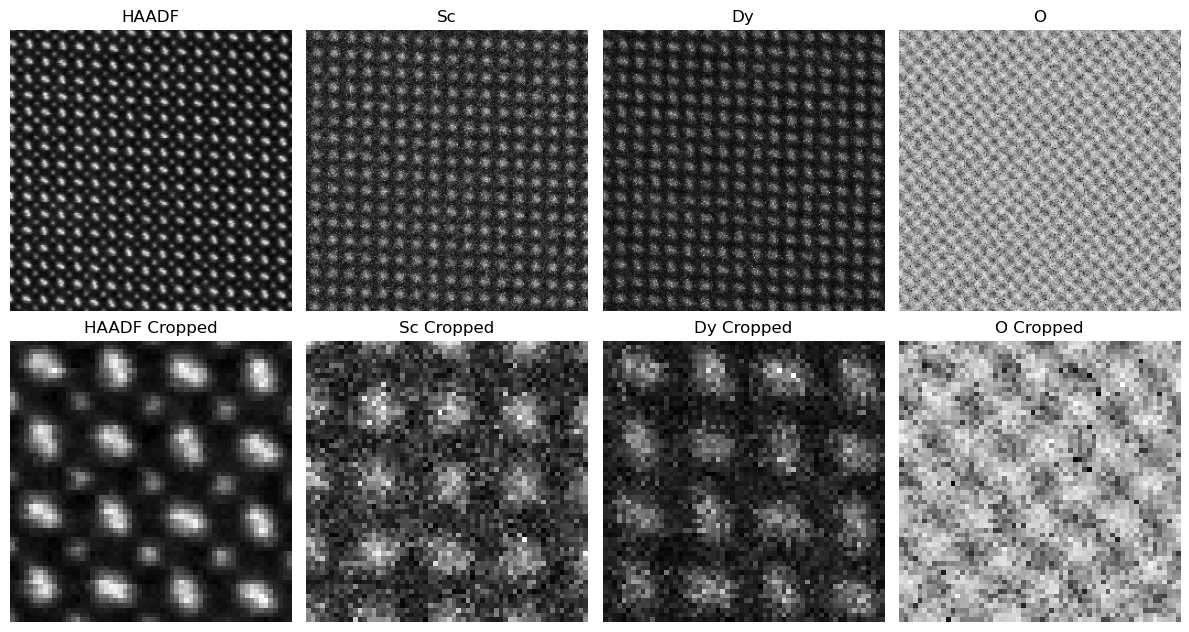
# Convergence Parameters
lambdaHAADF = 1/nz # Do not modify this
lambdaChem = 0.08
lambdaTV = 0.15; #Typically between 0.001 and 1
nIter = 30 # Typically 10-15 will suffice
bkg = 2.4e-1
# FGP TV Parameters
regularize = True; nIter_TV = 3; # xx represents the flattened 1D elastic maps we are trying to improve via the cost function
xx = xx0.copy()
# Auxiliary Functions for measuring the cost functions
lsqFun = lambda inData : 0.5 * np.linalg.norm(A.dot(inData**gamma) - HAADF) **2
poissonFun = lambda inData : np.sum(xx0 * np.log(inData + 1e-8) - inData)
# Main Loop
# Initialize the three cost functions components
costHAADF = np.zeros(nIter,dtype=np.float32); costChem = np.zeros(nIter, dtype=np.float32); costTV = np.zeros(nIter, dtype=np.float32);
for kk in tqdm(range(nIter)):
# Solve for the first two optimization functions $\Psi_1$ and $\Psi_2$
xx -= gamma * spdiags(xx**(gamma - 1), [0], nz*nx*ny, nz*nx*ny) * lambdaHAADF * A.transpose() * (A.dot(xx**gamma) - HAADF) + lambdaChem * (1 - xx0 / (xx + bkg))
# Enforce positivity constraint
xx[xx<0] = 0
# FGP Regularization if turned on
if regularize:
for zz in range(nz):
xx[zz*nPix:(zz+1)*nPix] = reg.fgp_tv( xx[zz*nPix:(zz+1)*nPix].reshape(nx,ny), lambdaTV, nIter_TV).flatten()
# Measure TV Cost Function
costTV[kk] += reg.tv( xx[zz*nPix:(zz+1)*nPix].reshape(nx,ny) )
# Measure $\Psi_1$ and $\Psi_2$ Cost Functions
costHAADF[kk] = lsqFun(xx); costChem[kk] = poissonFun(xx)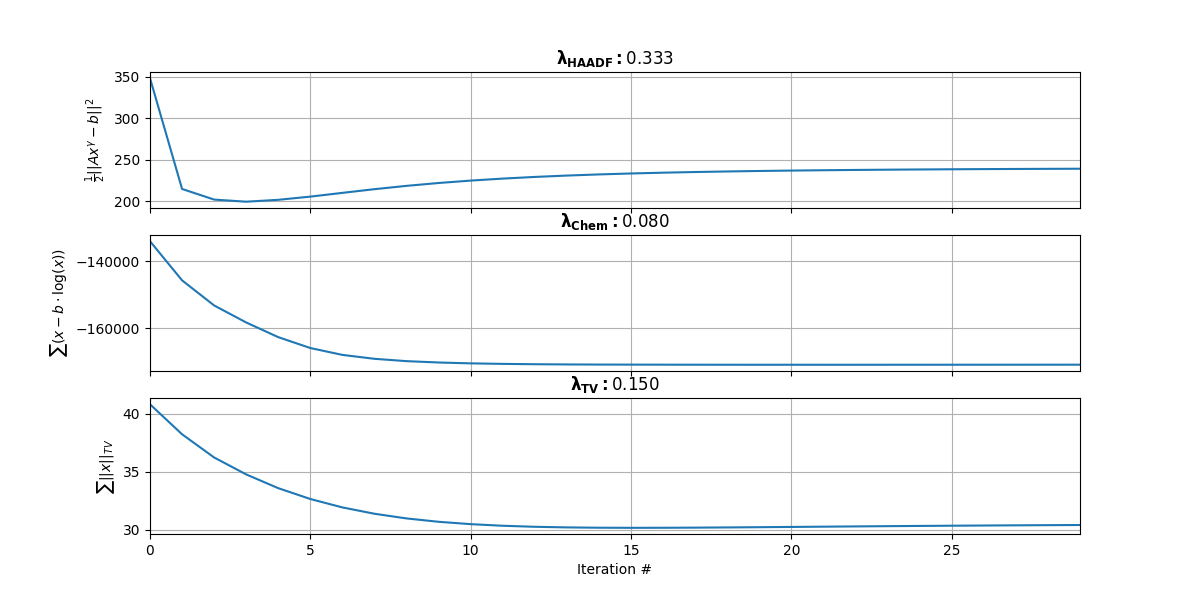
Figure 3.2:Convergence of 3 subparts of the multi-modal cost function. The top plot represents the first term that is dictated by . The middle plot represents the second term that is dictated by . The bottom plot represents the third TV term that is dictated by .
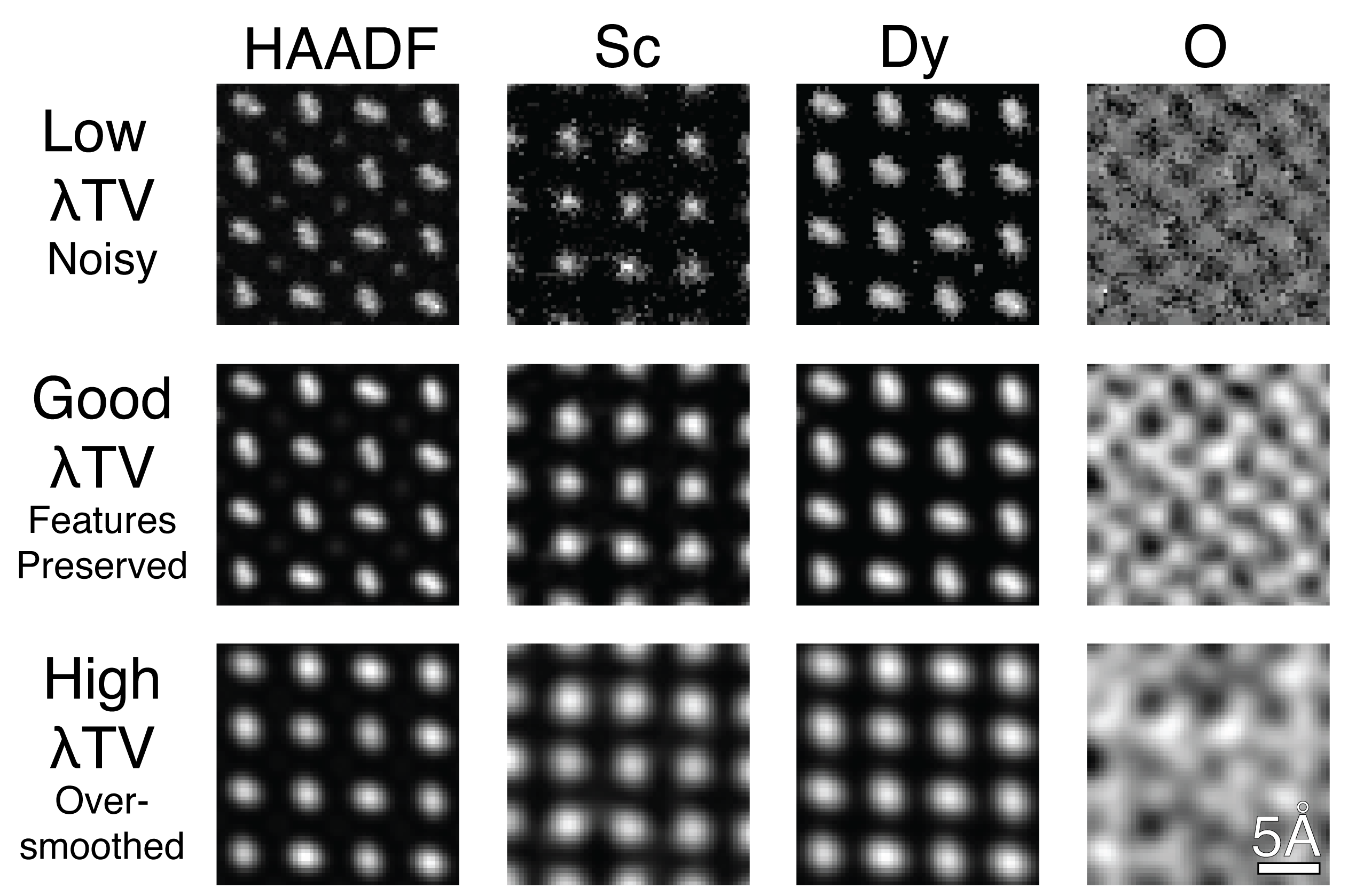
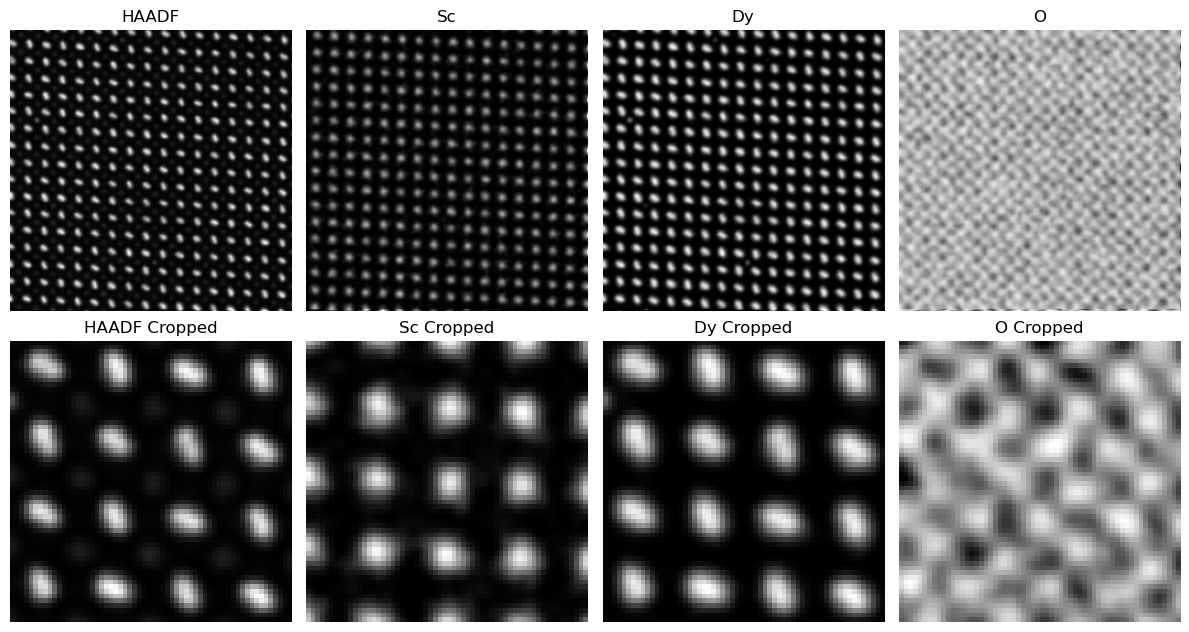
Figure 3.3:Comparison of TV weighting across different chemistries and HAADF. Too low of a TV results in noise and artifacts across images. Proper TV preserves fine features like the dumbell shape of the Dy particles, while reducing noise. High TV oversmoothes the image resulting in loss of important features for analysis.
# Display Cost Functions and Descent Parameters
utils.plot_convergence(costHAADF, lambdaHAADF, costChem, lambdaChem, costTV, lambdaTV)
# Show Reconstructed Signal
fig, ax = plt.subplots(2,len(elem_names)+1,figsize=(12,6.5))
ax = ax.flatten()
ax[0].imshow((A.dot(xx**gamma)).reshape(nx,ny),cmap='gray'); ax[0].set_title('HAADF'); ax[0].axis('off')
ax[1+len(elem_names)].imshow((A.dot(xx**gamma)).reshape(nx,ny)[70:130,25:85],cmap='gray'); ax[1+len(elem_names)].set_title('HAADF Cropped'); ax[1+len(elem_names)].axis('off')
for ii in range(len(elem_names)):
ax[ii+1].imshow(xx[ii*(nx*ny):(ii+1)*(nx*ny)].reshape(nx,ny),cmap='gray')
ax[ii+2+len(elem_names)].imshow(xx[ii*(nx*ny):(ii+1)*(nx*ny)].reshape(nx,ny)[70:130,25:85],cmap='gray')
ax[ii+1].set_title(elem_names[ii])
ax[ii+1].axis('off')
ax[ii+2+len(elem_names)].set_title(elem_names[ii]+' Cropped')
ax[ii+2+len(elem_names)].axis('off')
fig.tight_layout()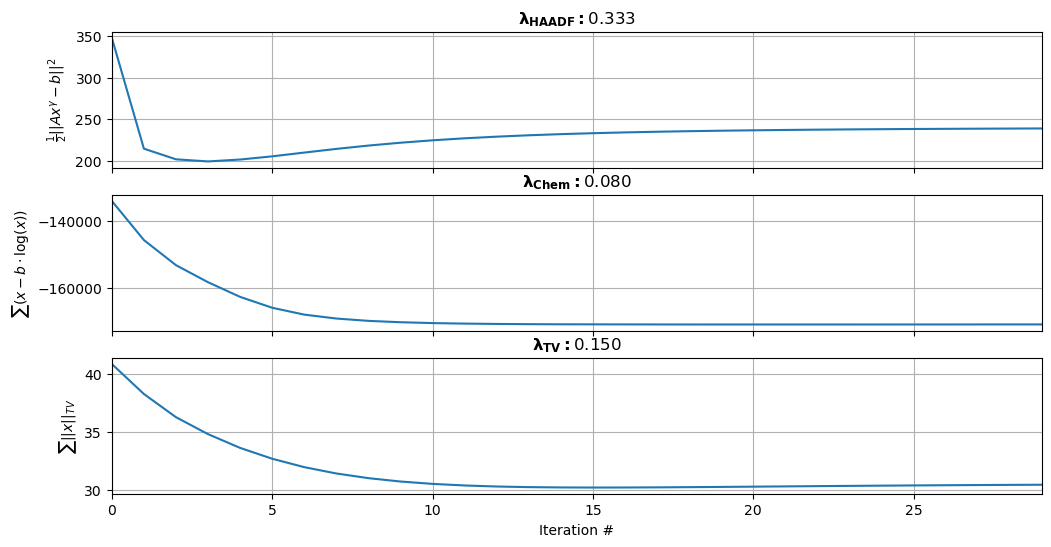
# Widgets for the parameters
kwargs = {
'style':{'description_width': 'initial'},
'layout':widgets.Layout(width='400px'),
'continuous_update': False,
'readout_format':'.3f'
}
lambdaChem_slider = widgets.FloatSlider(value=lambdaChem, min=0.001, max=1, step=0.001, description='lambdaChem',**kwargs)
lambdaTV_slider = widgets.FloatSlider(value=lambdaTV, min=0.001, max=1, step=0.001, description='lambdaTV',**kwargs)
nIter_slider = widgets.IntSlider(value=nIter, min=10, max=50, step=1, description='# Cost Function Iterations',**kwargs)
nIter_TV_slider = widgets.IntSlider(value=nIter_TV, min=1, max=10, step=1, description=' # TV Iterations',**kwargs)
def widget_wrapper(lambdaChem,lambdaTV,nIter,nIter_TV):
return_reconstruction_plots(
xx0,
HAADF,
A,
bkg,
(nx,ny,nz),
elem_names,
(70,130,25,85),
lambdaChem,
lambdaTV,
nIter,
nIter_TV
)
widgets.interact(widget_wrapper, lambdaChem=lambdaChem_slider, lambdaTV=lambdaTV_slider, nIter=nIter_slider, nIter_TV=nIter_TV_slider);#save_folder_name='test'
#utils.save_data(save_folder_name, xx0, xx, HAADF, A.dot(xx**gamma), elem_names, nx, ny, costHAADF, costChem, costTV, lambdaHAADF, lambdaChem, lambdaTV, gamma)
