Figures and Image Widgets
Figures are typically the most important part of any scientific paper, especially for studies focused on microscopy. We can include figures in many ways, for example by inserting conventional static figures using markdown:
Figure 1.1:HRTEM focal series reconstruction of a single-layer graphene GB. (a) Exit wave phase, enlarged in (b). (c) Exit wave amplitude, enlarged in (d). Adapted from Ophus et al. (2016).
However, figures such as Figure 1.1 are very limited. We need to make a trade-off between showing the entire FOV and zooming in to show the atomic details. This is solved in Figure 1.1 by having two panels for both the complex wave phase and amplitude, one showing the full FOV and another as an enlarged panel. These kinds of trade-offs are unavoidable for print journals where page space is limited.
However, since this journal is intended be read online, we can make more efficient use of the screen real estate. One method is to use tabbed viewing in order to show each figure panel individually:
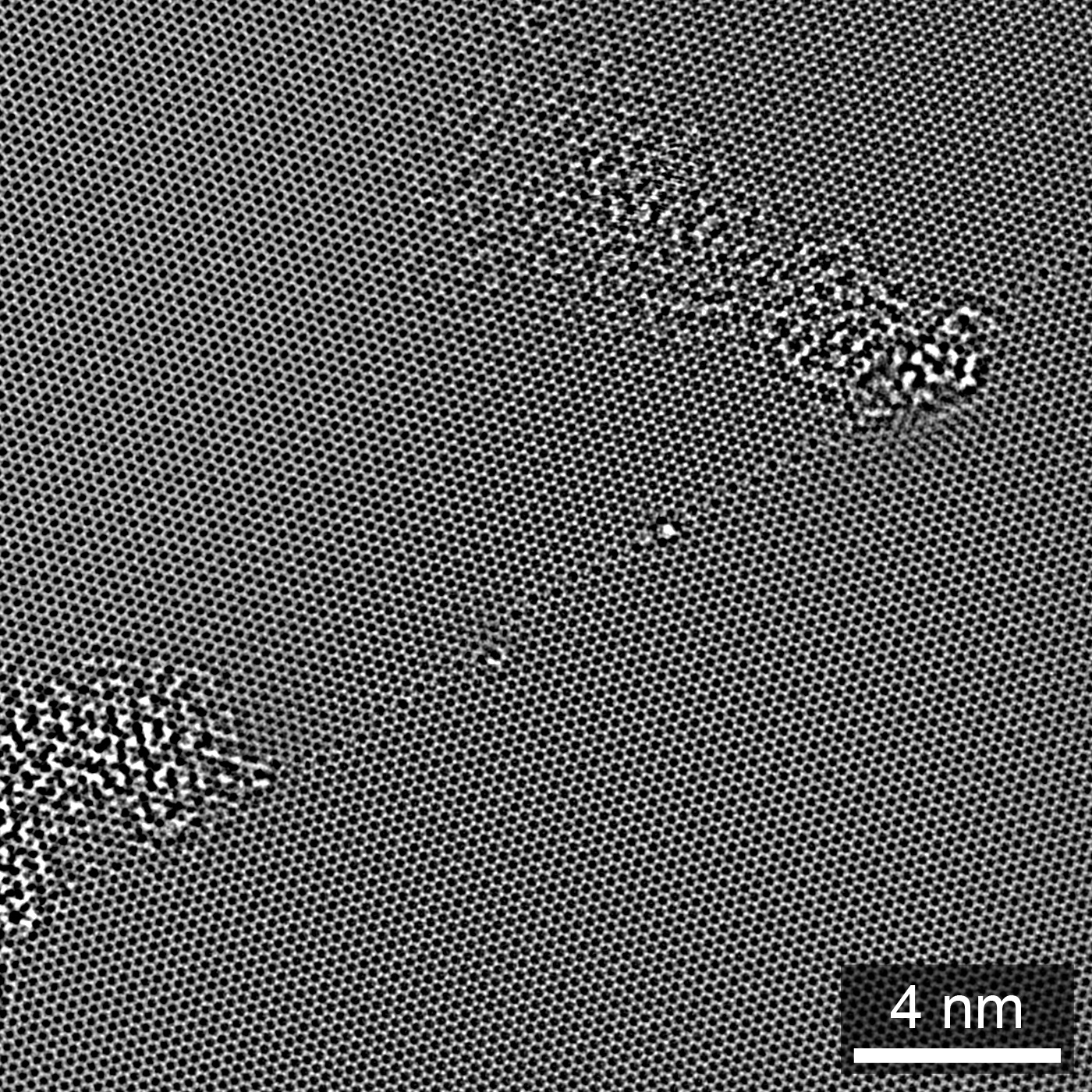
Figure 1.2:Exit wave phase of a single-layer graphene GB, from HRTEM focal series reconstruction. Adapted from Ophus et al. (2016).
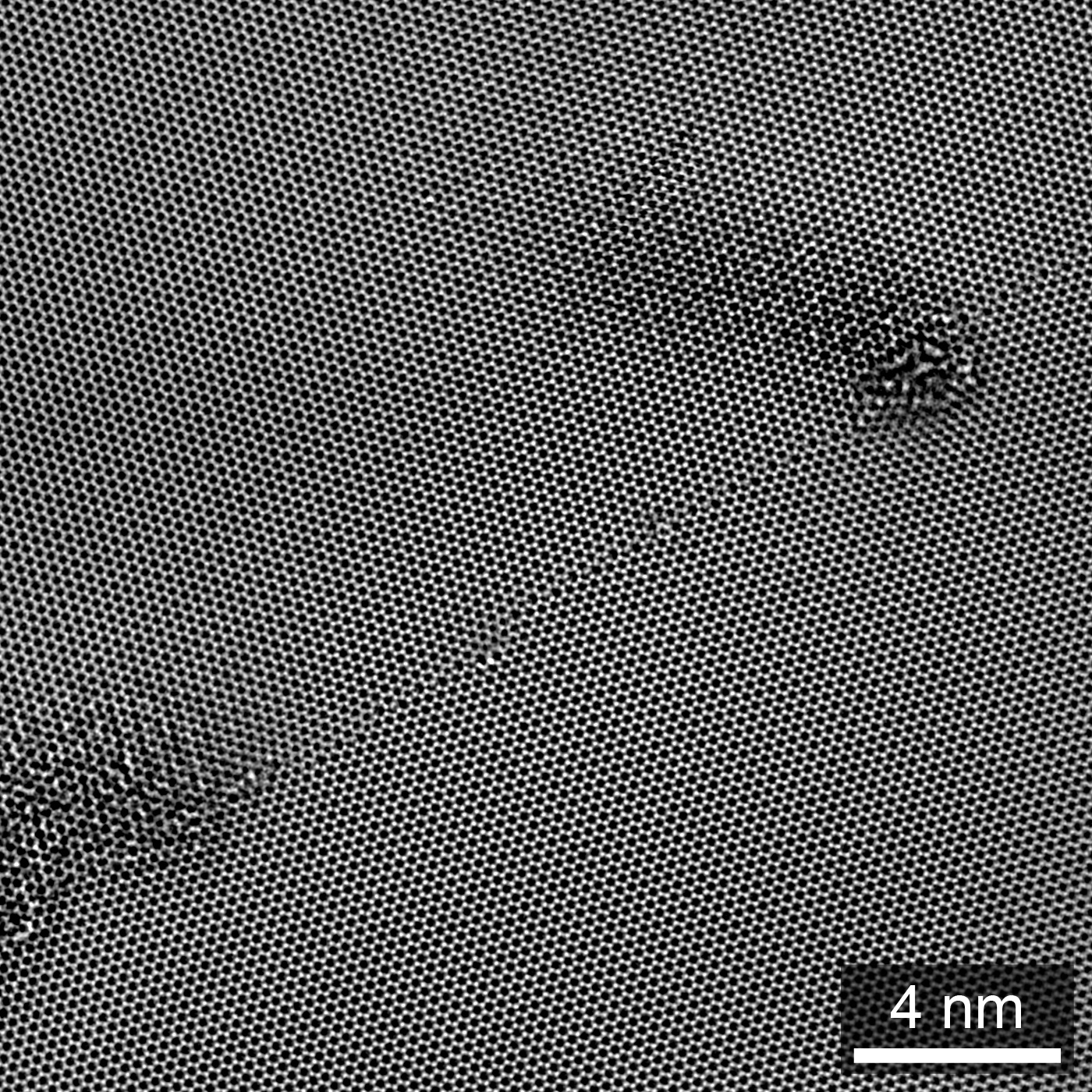
Figure 1.3:Exit wave amplitude of a single-layer graphene GB, from HRTEM focal series reconstruction. Adapted from Ophus et al. (2016).
Note that while we only display a single tab at a time, we can reference these panels individually: the EWR phase is shown in Figure 1.2, while the EWR amplitude is show in Figure 1.3. If you mouse over either of these links, previews of the figure panel will pop up.
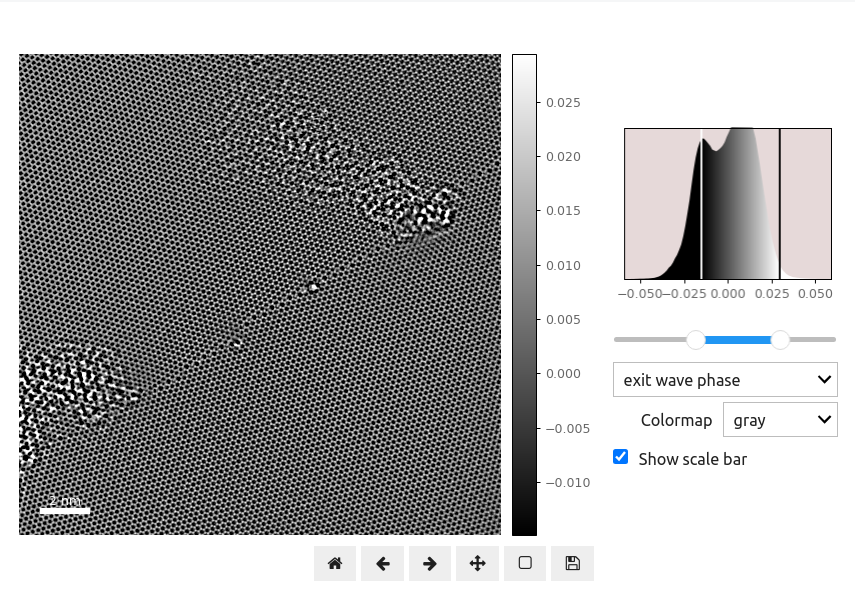
Tabbed image viewing is certainly more space-efficient than conventional figure panel layouts. However with python widgets we can do far better! In addition to placing multiple panels into the same space, we can also allow the reader to zoom in or out, pan around, and even change the image contrast. This is especially important for microscopy images which may have large dynamic ranges, or require both a large FOV and high resolution to fully appreciate.
Try playing around with this figure! You can zoom in, pan around, change the contrast range, and even change the colormap. Additionally we can use a dropdown menu to select either the EWR phase or amplitude. We reference Figure 1.4 in the same manner as before.
- Ophus, C., Rasool, H. I., Linck, M., Zettl, A., & Ciston, J. (2016). Automatic software correction of residual aberrations in reconstructed HRTEM exit waves of crystalline samples. Advanced Structural and Chemical Imaging, 2, 1–10. https://doi.org/10.1186/s40679-016-0030-1